
Inadequate support: The article cites no data or information to support its claims, which are based solely on hearsay from an anonymous person on a “medical forum”.

BEHAUPTUNG: Here’s the problem, we are testing people for any strain of a Coronavirus. Not specifically for COVID-19. There are no reliable tests for a specific COVID-19 virus.
SUMMARY
This claim appeared in an article published on GlobalResearch.ca by Julian Rose, which was later propagated in a YouTube video by Jerry Day. The article has received more than 11,000 engagements on Facebook, while the video has been viewed more than 750,000 times. The claims are reported by the article as originating from “a widely respected professional scientist in the US”, which were written in a “medical forum”, although the article does not name this scientist.
The article criticizes the laboratory technique known as PCR, or polymerase chain reaction. This is a widely-used technique in laboratories worldwide and was invented by Kary Mullis, who received the 1993 Nobel Prize in Chemistry for this work. The article is correct in stating that PCR is used to make many copies of a particular segment of nucleic acid (DNA/RNA), a process known as amplification, enabling scientists to detect tiny amounts of nucleic acid in a sample.
However, the article errs in stating that “PCR basically takes a sample of your cells and amplifies any DNA to look for ‘viral sequences’.” PCR tests do not indiscriminately amplify any DNA in a sample; they are designed to target only the nucleic acid sequence from the microorganism of interest (specificity), in this case SARS-CoV-2, the virus causing COVID-19.
PCR specificity is achieved through proper design of primers, which are short nucleic acid sequences designed to bind or anneal only to the target, and this is made possible through the rule of complementary base pairing. Bases, or nucleotides, are the individual building blocks of DNA and RNA sequences, and each base pairs with only one other base. In DNA for example, adenosine pairs only to thymine, and cytosine pairs only to guanine.
There are many requirements involved in designing appropriate PCR primers and this article by the Diamantina Institute of the University of Queensland outlines several of them and why they are important to consider:
“A lot of work has to go into designing the primers. Firstly, we need to know the sequence of the section of DNA we are wanting to amplify, particularly the ‘beginning’ (5’) and ‘ending’ (3’) of the sequence. The primers need to be designed so that they are complementary to a unique sequence of nucleotides ‘upstream’ and ‘downstream’ of the sequence of interest. They cannot match a sequence within the area of interest (or the PCR will start off too late and miss a portion of the area we want to amplify), and they should also not have complementary regions within themselves (or they will fold over and bind to themselves, forming a “hairpin”. Lastly, the forward and reverse primers should not be complementary, or they will anneal to each other and form a ‘primer dimer’.”
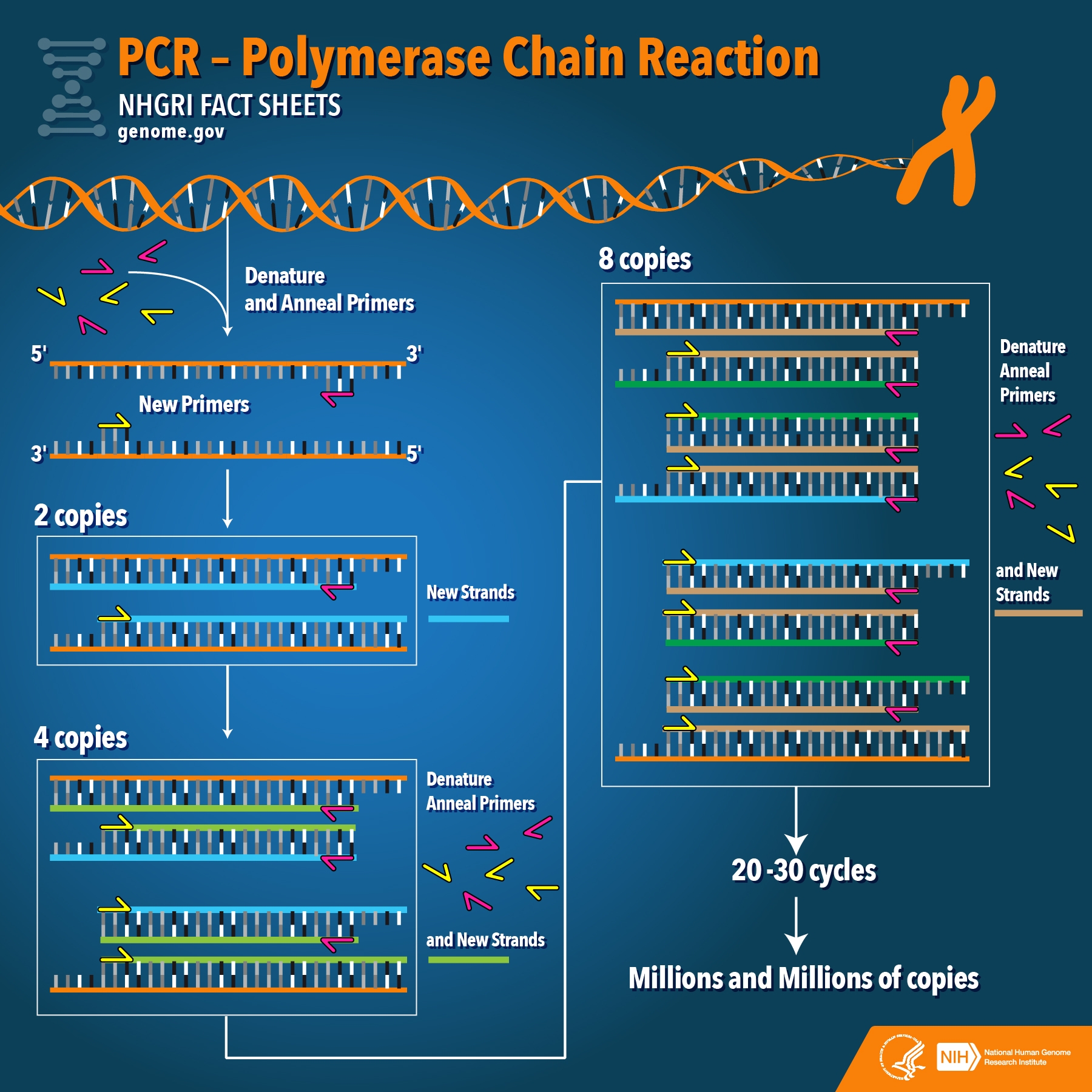
Figure—A graphic by the U.S. National Human Genome Research Institute showing how PCR works.
Thanks to the efforts of scientists, we now know the full genome of SARS-CoV-2[1], which makes it possible to design specific primers for the virus that recognize unique segments of nucleic acid sequences and do not anneal to those found in other coronaviruses. Furthermore, primer annealing to the target sequence is necessary to start PCR amplification. If the primers do not match a nucleic acid sequence—for example in the case of a different coronavirus—then PCR amplification of that mismatched sequence cannot occur. In short, the claim that the test for COVID-19 detects “any strain of coronavirus” is false and unfounded.
The article also wrongly claims that PCR is “useless at telling you how much virus you may have”. On the contrary, variants of PCR, such as quantitative PCR, have been used for measuring viral load long before the COVID-19 pandemic occurred[2,3,4]. Viral load is generally related to transmissibility and severity of infection and therefore clinically important. For example, viral load testing is commonly done for HIV patients in order to manage their condition. In this article by the World Economic Forum, David Duong, Director of the Harvard Medical School Program in Global Primary Care and Social Change, stated that most countries—including the U.S.—are using reverse transcription PCR (RT-PCR) for COVID-19. RT-PCR is a variant of PCR that is able to detect RNA (SARS-CoV-2 is an RNA virus).
In summary, the article’s claims that PCR indiscriminately detects any coronavirus and that PCR cannot measure viral load does not hold up under scrutiny. Overall, this is yet another article propagating the unfounded conspiracy theory that COVID-19 is a pandemic “manufactured” to create panic and that it is not actually a new virus—numerous studies have recognized SARS-CoV-2 as a novel coronavirus not previously identified, showing this claim to be false[5,6].
SCIENTISTS‘ FEEDBACK
It’s false. SARS-CoV-2, the agent of COVID-19, has unique sequences which are different from other coronaviruses. The PCR tests are very specific.
REFERENCES
- 1 – Zhou et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature.
- 2 – Mackay et al. (2002) Real-time PCR in virology. Nucleic Acids Research.
- 3 – Watzinger et al. (2004) Real-Time Quantitative PCR Assays for Detection and Monitoring of Pathogenic Human Viruses in Immunosuppressed Pediatric Patients. Journal of Clinical Microbiology.
- 4 – Engelmann et al. (2008) Rapid quantitative PCR assays for the simultaneous detection of herpes simplex virus, varicella zoster virus, cytomegalovirus, Epstein-Barr virus, and human herpesvirus 6 DNA in blood and other clinical specimens. Journal of Medical Virology.
- 5 – Zhu et al. (2020) A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England Journal of Medicine.
- 6 – Lu et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet.


