
Flawed reasoning: The genetic sequence used as template by the researchers from the Wuhan Institute of Virology and the monkeypox virus responsible for the 2022 outbreak come from two, genetically distinct, monkeypox subgroups. Suggestions that the published research from the Wuhan Institute of Virology could be linked to the 2022 monkeypox outbreak are thus baseless.

RECLAMACION: Researchers from the Wuhan Institute of Virology (WIV) “assembled monkeypox strains” using techniques “flagged for potentially creating a ‘contagious pathogen’”; it could be linked to the 2022 monkeypox outbreak
REVIEW
Reports of unusual monkeypox cases hit the headlines in May 2022, with at least 257 cases and zero deaths in 23 countries as of 30 May 2022. Monkeypox is a viral disease caused by the monkeypox virus, a member of the Orthopoxvirus genus, which also includes the now-eradicated variola virus responsible for smallpox. The disease is characterized by fever, swelling of the lymph nodes, and muscle pain, followed by rashes.
Monkeypox is rare and mostly occurs in Central and West Africa. While some outbreaks happened in other regions, they were usually associated with people who had traveled to regions where the virus circulates or with imported animals. However, the large majority of cases of the 2022 outbreak occurred among people with no travel records to regions where the disease is endemic, which drew attention of authorities, news outlets and social media alike.
The National Pulse website published an article claiming that researchers from the Wuhan Institute of Virology (WIV) “assembled monkeypox strains” a couple of months before the onset of the 2022 outbreak, using techniques “flagged for potentially creating a ‘contagious pathogen’”. These claims were also repeated by American radio host Dan Bongino.
The WIV became famous during the COVID-19 pandemic as it is a renowned center for coronavirus research and has been the target of many claims that SARS-CoV-2 had leaked from or was engineered there. These claims are still unsupported by scientific evidence to date. Similarly, the monkeypox strain assembly claim is inaccurate, as we show below.
It is based on a study published in February 2022 by Yang et al.[1]. The aim of this research was to test improvements of an already established genetic technique called transformation-associated recombination (TAR). TAR allows the assembly of long DNA segments with a higher rate of success than alternative techniques[2]. It is therefore widely used in several fields such as genomics or synthetic biology.
Over the years, several improvements were made to the original TAR method in order to improve its success rate. One such technique had previously been described in 2003 for the assembly of DNA sequence of the size of a gene, but hadn’t been tested for their feasibility in assembling longer DNA segments[3]. This is what Yang et al. set out to test, using the monkeypox virus genome as a model of study.
However, the claim that the WIV researchers “assembled monkeypox strains” is inaccurate. This is because Yang et al. only assembled a limited portion of the monkeypox genome, specifically less than one-third of the full genome, or 55,000 base pairs out of approximately 200,000 base pairs[4]. The number of base pairs in a genome tells us the size of a genome (see the illustration below). Therefore, their work didn’t produce full viruses.
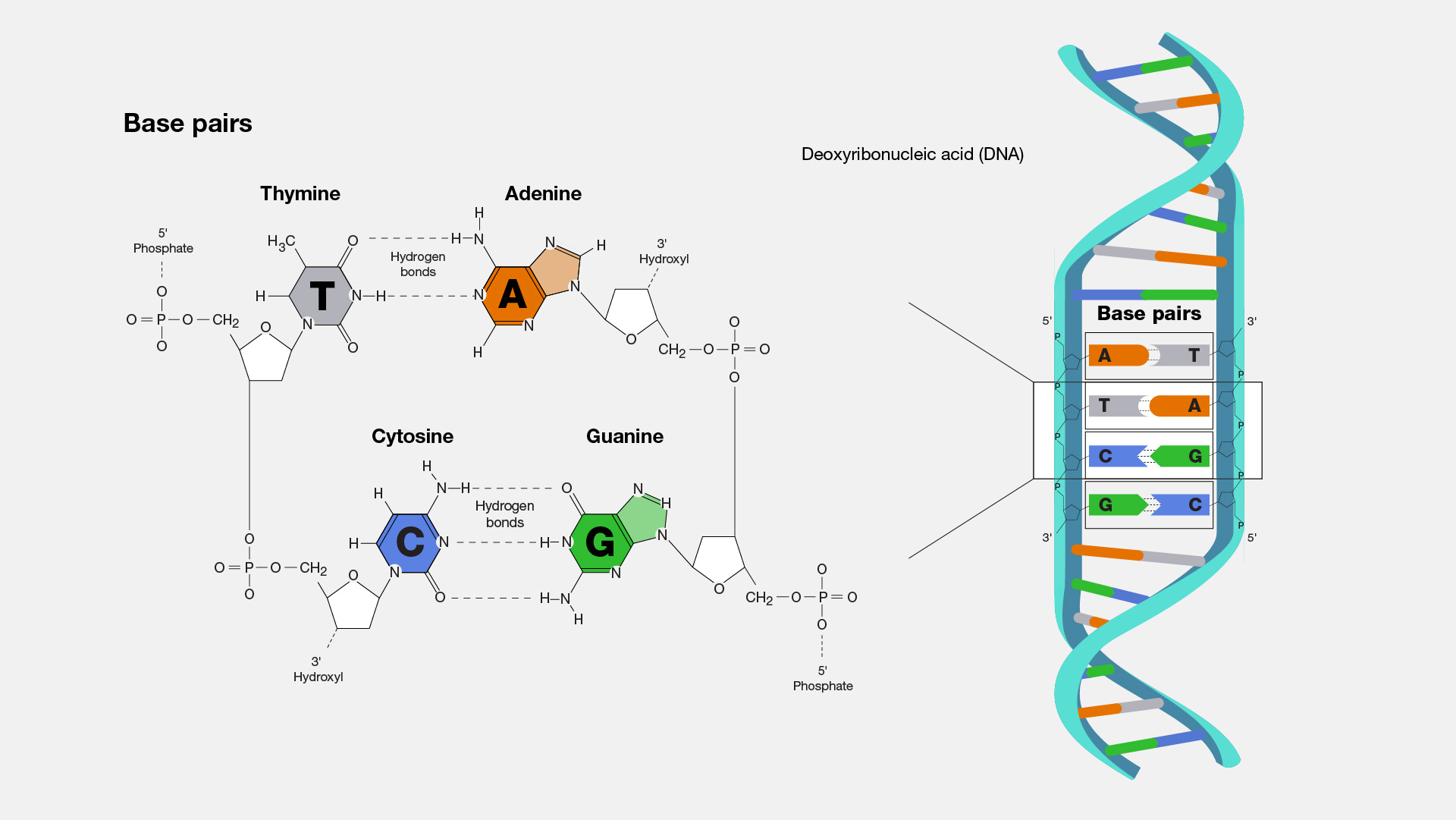
This illustration shows the different building blocks of DNA (called bases or nucleotides) and their respective pairings. Adenine always pairs with thymine, while cytosine always pairs with guanine. The number of base pairs that make up a genome is also a measure of the size of the genome. Source: U.S. National Human Genome Research Institute.
In fact, the researchers had limited their work to a fragment of the monkeypox genome specifically out of concerns for safety, which is clearly stated in the study:
«In this study, although a full-length viral genome would be the ideal reference template for detecting [the monkeypox virus] by qPCR, we only sought to assemble a 55 [thousands base pairs] viral fragment, less than one-third of the [monkeypox] genome. This assembly product is fail-safe by virtually eliminating any risk of recovering into an infectious virus.«
It is also misleading to suggest that the technique Yang et al. used was “flagged for potentially creating a ‘contagious pathogen’”—suggesting that the only purpose of using TAR is to assemble an infectious virus. Indeed, the technique alone cannot create a contagious pathogen. It ultimately depends on how researchers use the technique.
For instance, a similar technique was used in 2022 to fully synthesize an infectious SARS-CoV-2 virus by a Swiss team[5]. In 2018, Canadian researchers synthesized a whole, infectious horsepox virus[6]. In both cases, the research teams purposely aimed to reconstitute a whole, functional virus by using the whole genome as target DNA sequence to be assembled. But this is not what Yang et al. did. In fact, they specifically avoided assembling the whole monkeypox genome.
Claiming that the TAR technique was “flagged for potentially creating ‘contagious viruses’” would be as misleading as claiming that soldering was flagged for potentially creating weapons of mass destruction, merely because it is used at some point in the process of making weapons .
Finally, the National Pulse and Bongino also hinted that the WIV research might be linked to the current 2022 monkeypox outbreak. Indeed, the National Pulse concluded its article by claiming that WIV had performed similar research on coronavirus in spite of ongoing safety issues at the laboratory, allegedly. In his video, Bongino associated the confirmed cases of monkeypox in the U.S. with the inaccurate claim that monkeypox strains were assembled by WIV. By juxtaposing these ideas, the article and the video conveyed the baseless idea that the 2022 monkeypox outbreak could be linked to WIV research.
However, this idea is scientifically unfounded, as shown by genetic differences between the monkeypox virus from the 2022 outbreak and the monkeypox genome fragment used by Yang et al. Monkeypox viruses can be divided into two subgroups, or clades, based on genetic differences between them: the Congo Basin clade, and the West Africa clade[7]. Yang et al. used the monkeypox genome Congo_2003_358, which is from the Congo Basin clade[1,7]. However, the current 2022 outbreak is caused by a monkeypox strain belonging to the West Africa clade.
In other words, the genome used by WIV researchers and the monkeypox virus circulating during the 2022 outbreak come from two, genetically distinct groups of monkeypox virus, invalidating the allegations that the published WIV research is somehow tied to the 2022 outbreak.
In summary, no monkeypox virus was ever assembled in the WIV study cited by the National Pulse and Bongino. The research team only assembled a third of the entire monkeypox genome, whereas the entire virus’ genetic sequence would be necessary to produce a functional virus. Furthermore, the monkeypox genome used in the study as a model and the genome from the virus circulating in 2022 are genetically distinct. The 2022 monkeypox outbreak is thus unrelated to the study published by WIV researchers.
REFERENCES
- 1 – Yan et al. (2022) Efficient assembly of a large fragment of monkeypox virus genome as a qPCR template using dual-selection based transformation-associated recombination. Virologica Sinica.
- 2 – Kouprina & Larionov (2016) Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma.
- 3 – Noskov et al. (2003) A general cloning system to selectively isolate any eukaryotic or prokaryotic genomic region in yeast. BMC Genomics.
- 4 – Zhao et al. (2016) Finishing monkeypox genomes from short reads: assembly analysis and a neural network method. BMC Genomics.
- 5 – Thao et al. (2020) Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature.
- 6 – Noyce et al. (2018) Construction of an infectious horsepox virus vaccine from chemically synthesized DNA fragments. PLOS One.
- 7 – Likos et al. (2005) A tale of two clades: monkeypox viruses. Journal of General Virology.


